DeRIP2 Command Line Interface
Basic usage
For aligned sequences in 'mintest.fa':
- Any column with >= 70% gap positions will not be corrected and a gap inserted in corrected sequence.
- Bases in column must be >= 80% C/T or G/A
- At least 50% bases in a column must be in RIP dinucleotide context (C/T as CpA / TpA) for correction.
- Default: Inherit all remaining uncorrected positions from the least RIP'd sequence.
- Mask all substrate and product motifs from corrected columns as ambiguous bases (i.e. CpA to TpA --> YpA)
Basic usage with masking
derip2 -i tests/data/mintest.fa \
--max-gaps 0.7 \
--max-snp-noise 0.2 \
--min-rip-like 0.5 \
--mask \
-d results \
--prefix derip_output
Output:
results/derip_output.fasta- Corrected sequenceresults/derip_output_alignment.fasta- Alignment with masked correctionsresults/derip_output_masked_alignment.fasta- Alignment with masked corrections
With vizualization
The --plot option will create a visualization of the alignment with RIP markup. The --plot-rip-type option can be used to specify the type of RIP events to be displayed in the alignment visualization product, substrate, or both.
derip2 -i tests/data/mintest.fa \
--max-gaps 0.7 \
--max-snp-noise 0.2 \
--min-rip-like 0.5 \
--plot \
--plot-rip-type both \
-d results \
--prefix derip_output
Output:
results/derip_output.fasta- Corrected sequenceresults/derip_output_masked_alignment.fasta- Alignment with masked correctionsresults/derip_output_visualization.png- Visualization of the alignment with RIP markup
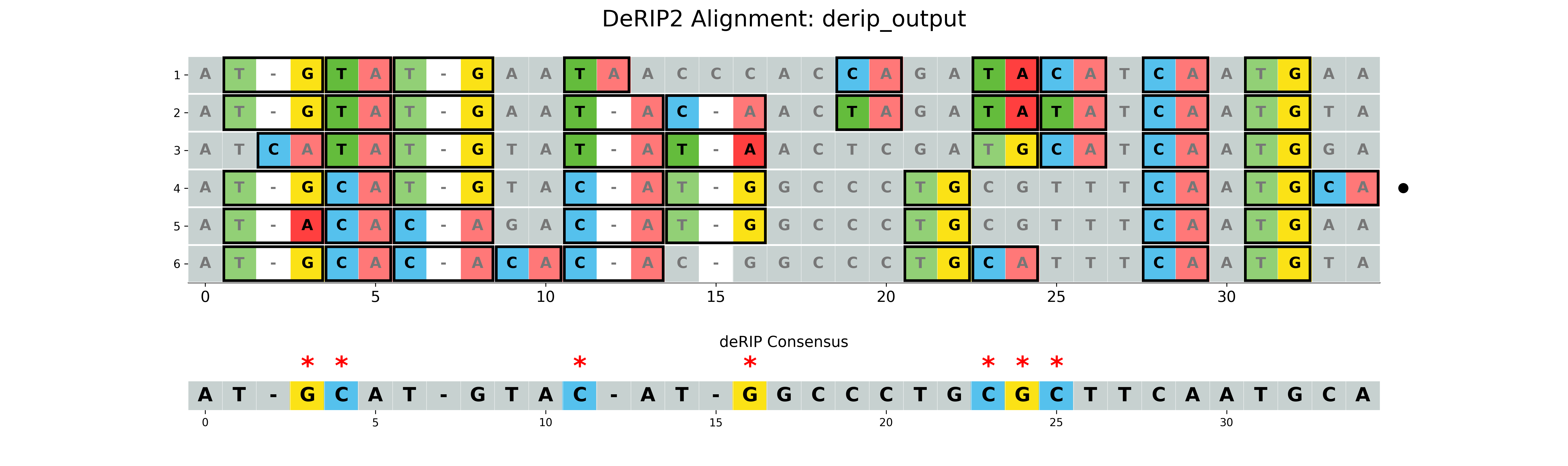
Using maximum GC content for filling
By default uncorrected positions in the output sequence are filled from the sequence with the lowest RIP count. If the --fill-max-gc option is set, remaining positions are filled from the sequence with the highest G/C content sequence instead.
derip2 -i tests/data/mintest.fa \
--max-gaps 0.7 \
--max-snp-noise 0.2 \
--min-rip-like 0.5 \
--fill-max-gc \
-d results \
--prefix derip_gc_filled
Alternatively, the --fill-index option can be used to force selection of alignment row to fill uncorrected positions from by row index number (indexed from 0). Note: This will override the --fill-max-gc option.
Correcting all deamination events
If the --reaminate option is set, all deamination events will be corrected, regardless of RIP context.
--plot-rip-type product is used to highlight the product of RIP events in the visualization.
Non-RIP deamination events are also highlighted.
derip2 -i tests/data/mintest.fa \
--max-gaps 0.7 \
--reaminate \
-d results \
--plot \
--plot-rip-type product \
--prefix derip_reaminated
Output:
results/derip_reaminated.fasta- Corrected sequence using highest GC content sequence for fillingresults/derip_reaminated_alignment.fasta- Alignment with corrected sequence appendedresults/derip_reaminated_vizualization.png- Visualization of the alignment with RIP markup
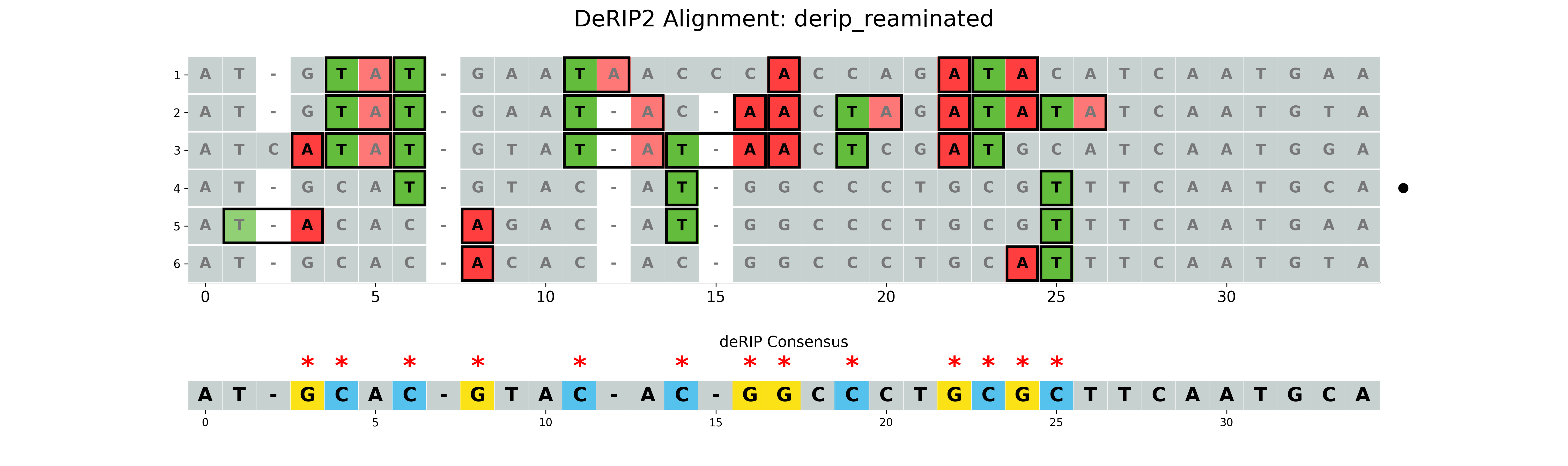
Standard Options
--version Show the version and exit.
-i, --input TEXT Multiple sequence alignment. [required]
-g, --max-gaps FLOAT Maximum proportion of gapped positions in
column to be tolerated before forcing a gap
in final deRIP sequence. [default: 0.7]
-a, --reaminate Correct all deamination events independent
of RIP context.
--max-snp-noise FLOAT Maximum proportion of conflicting SNPs
permitted before excluding column from
RIP/deamination assessment. i.e. By default
a column with >= 0.5 'C/T' bases will have
'TpA' positions logged as RIP events.
[default: 0.5]
--min-rip-like FLOAT Minimum proportion of deamination events in
RIP context (5' CpA 3' --> 5' TpA 3')
required for column to deRIP'd in final
sequence. Note: If 'reaminate' option is set
all deamination events will be corrected.
[default: 0.1]
--fill-max-gc By default uncorrected positions in the
output sequence are filled from the sequence
with the lowest RIP count. If this option is
set remaining positions are filled from the
sequence with the highest G/C content.
--fill-index INTEGER Force selection of alignment row to fill
uncorrected positions from by row index
number (indexed from 0). Note: Will override
'--fill-max-gc' option.
--mask Mask corrected positions in alignment with
degenerate IUPAC codes.
--no-append If set, do not append deRIP'd sequence to
output alignment.
-d, --out-dir TEXT Directory for deRIP'd sequence files to be
written to.
-p, --prefix TEXT Prefix for output files. Output files will
be named prefix.fasta,
prefix_alignment.fasta, etc. [default:
deRIPseq]
--plot Create a visualization of the alignment with
RIP markup.
--plot-rip-type [both|product|substrate]
Specify the type of RIP events to be
displayed in the alignment visualization.
[default: both]
--loglevel [DEBUG|INFO|WARNING|ERROR|CRITICAL]
Set logging level. [default: INFO]
--logfile TEXT Log file path.
-h, --help Show this message and exit.